Levels of Protein Structure
The structure of a protein is very significant to its function. To understand how a protein molecule forms its final conformation, we need to understand the four structural levels of proteins: primary structure, secondary structure, tertiary structure, and quaternary structure.
Primary Structure
The primary structure of a protein refers to the order of amino acid residues in the polypeptide chain of the protein. The amino acid is an organic molecule that binds to other amino acids by the peptide bond to form a peptide chain. Amino acids in the peptide chain are called residues. Although hundreds of amino acids are found in nature, proteins are made of 20 kinds of amino acids. Based on the properties of the free radicals at the terminus, the ends of the polypeptide chain are referred to as the amino terminus (N-terminus) and the carboxy terminus (C-terminus), respectively. The residue count always starts from the N-terminus. The amino acid sequence is unique to the protein and is the basis for understanding its structure and function. And the sequence can be determined by methods such as Edman degradation and mass spectrometry.
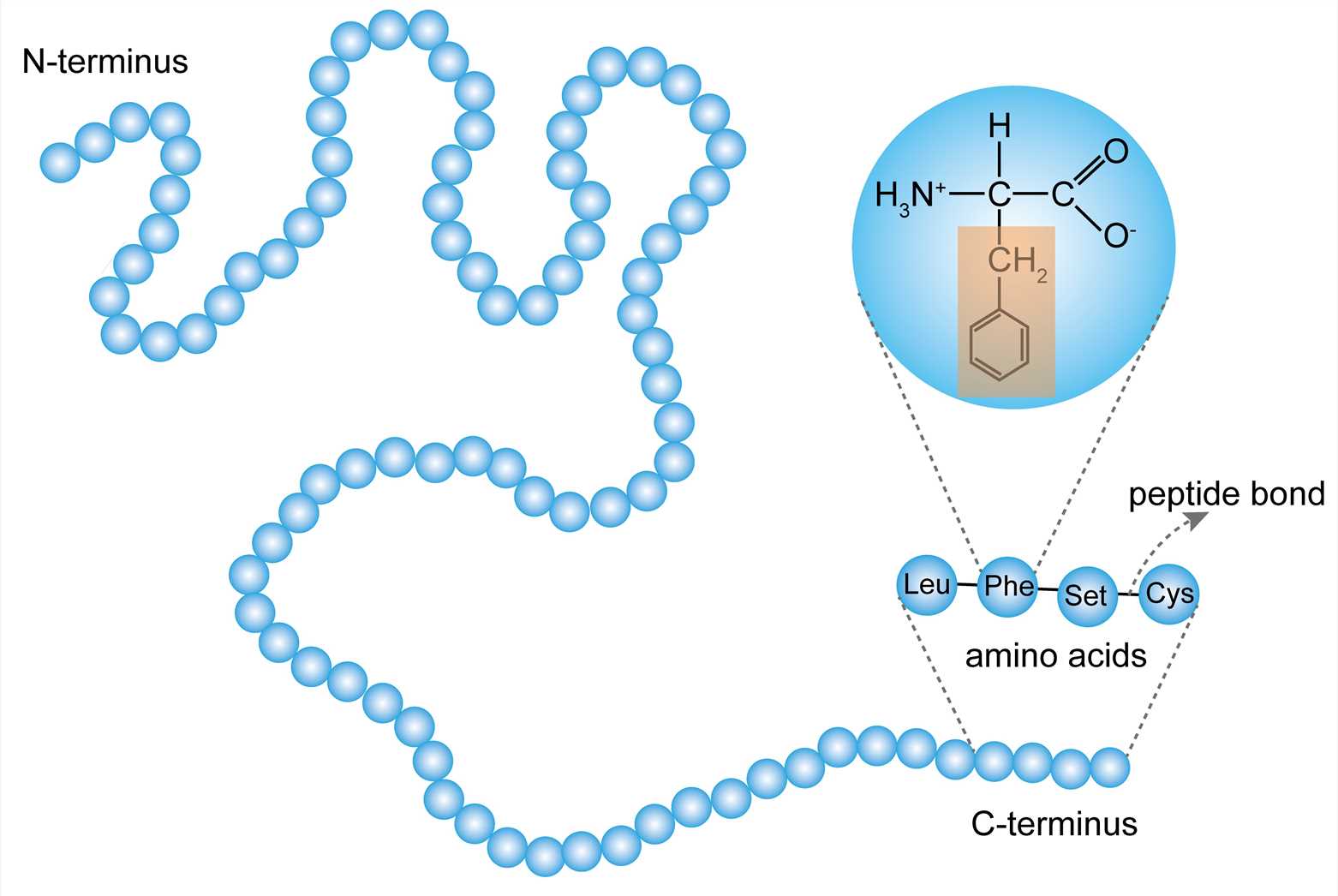 Figure 1. The primary structure.
Figure 1. The primary structure.
- Gene sequence determines the primary structure
The primary structure of a protein is determined by the order in which the genetic code is assigned to the corresponding gene of the protein. The specific nucleotide sequence in DNA is transcribed to mRNA, and the genetic code is read by the ribosome during translation. Various amino acids are bound together in the order of the genetic code to generate a peptide chain. Therefore, the genetic code can be used to directly acquire the corresponding amino acid sequence from the gene sequence. However, post-translational modifications such as disulfide bond formation, phosphorylation, and glycosylation, which are also generally considered to be part of the primary structure, cannot be obtained from the gene sequence.
- Primary structure determines the high-level structure
Each of the 20 amino acids constituting the protein has its own side chain, and the physicochemical properties and spatial arrangement of the side chain groups are different. When they are combined in a different order, a variety of spatial structures can be formed, therefore, the primary structure of a protein determines its secondary, tertiary, and other high-level structures. However, the polypeptide chains of proteins don’t extend linearly, but rather fold and coil to form a unique stable spatial structure. Hence, it is not possible to fully understand the biological activity and physicochemical characteristics of proteins merely by measuring their amino acid composition and amino acid sequence.
Secondary Structure
The secondary structure of a protein refers to the partial space arrangement of the peptide backbone atoms.
- α-helix and β-sheet
The most basic secondary structure types α-helix and β-sheet were proposed in 1951. Both structures are maintained by hydrogen bonds formed between the main-chain peptide groups. They have a regular geometry, being constrained to specific values of the dihedral angles ψ and φ on the Ramachandran plot.
Each helix of the α-helix structure contains 3.6 amino acid residues with a pitch of 0.54 nm. All peptide bonds in the α-helix structure participate in the formation of hydrogen bonds to maintain the stability of the helix.
The β-sheet structure consists of β-strands which can be arranged in parallel or antiparallel patterns. The adjacent peptide chains or peptide fragments are connected by hydrogen bonds to form a sheet structure.
The portion of the protein structure that has no regular conformational angle is called the loop or coil region, which often appears on the surface of proteins.
- Supersecondary structure
Several sequential secondary structures may form a "supersecondary structure", such as a combination of alpha helixes (αα), a combination of β-sheets (βββ), and a combination of alpha helixes and β-sheets (βαβ). The supersecondary structures act as building blocks for tertiary structures in a variety of proteins.
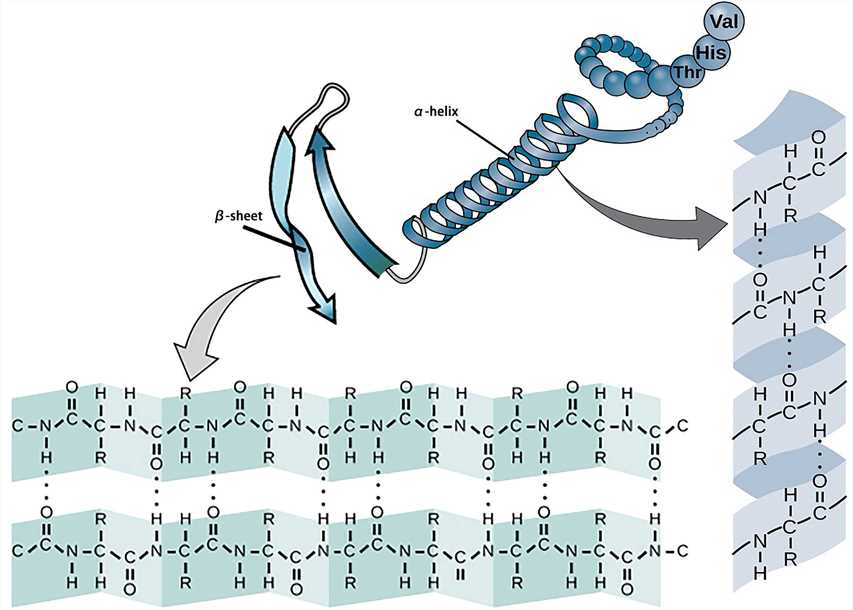 Figure 2. The secondary structure.
Figure 2. The secondary structure.
Tertiary Structure
The polypeptide chain of a protein further folds and curls based on various secondary structures to form a regular three-dimensional space structure, which is called the protein’s tertiary structure.
- The interaction between the amino acid side chains
The tertiary structure is mainly due to the interaction between the side chains of the amino acids constituting the protein. The stability of the tertiary structure mainly depends on secondary bonds, including hydrogen bonds, hydrophobic bonds, ionic bonds, van der Waals forces, and so on. These secondary bonds can be found between the R groups of residues whose primary structure numbers are far apart, thus the tertiary structure mainly refers to the binding between the side chains of amino acid residues.
The secondary bonds are non-covalent bonds, which are susceptible to changes in pH, temperature, ionic strength, etc. The disulfide bond doesn't belong to secondary bonds, but it is possible to link the two distant peptides together, which plays an important role in the stability of the tertiary structure.
- Protein domain
It is also considered that the tertiary structure refers to the formation of side-chain conformation based on the conformation of the peptide backbone. The side chain conformation is primarily formed into the so-called protein domain, which is a region of a tightly globular structure that is separated in the structure of a protein subunit. Additionally, the domain is usually a protein structure surrounding a single hydrophobic core.
Many domains are not unique to the protein products of a gene or a gene family but appear in a variety of proteins. They predominate in the biological function of the proteins because of their independence and stability, so chimeric proteins can be prepared by exchanging domains between one protein and another through genetic engineering technology.
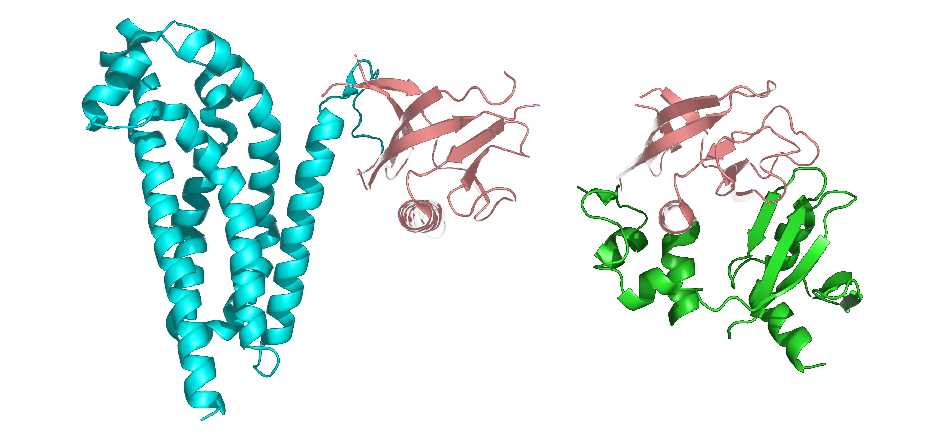 Figure 3. The representation of protein domains. The two shown protein structures share a common domain (carnation).
Figure 3. The representation of protein domains. The two shown protein structures share a common domain (carnation).
- Fibrous protein and globular protein
Proteins with a tertiary structure can be classified into fibrous proteins and globular proteins according to their appearance. The fibrous protein is slender (the long axis is more than 10 times longer than the short axis), such as fibroin; the globular protein is substantially spherical (the length of the long axis is similar to that of the short axis), such as plasma albumin, globulin, and myoglobin.
The globular protein forms hydrophilic and hydrophobic regions. The hydrophilic region is mostly on the surface of protein molecules and consists of many hydrophilic side chains. The hydrophobic region which is composed of hydrophobic side chains is mostly in the inner part of molecules. The hydrophobic region often forms “protein cavities” or “protein pockets”, and some prosthetic groups are embedded in them to become active sites.
In most cases, proteins need to form a tertiary structure to perform their specific biological functions, therefore, correct protein folding is an important consideration in biotechnology.
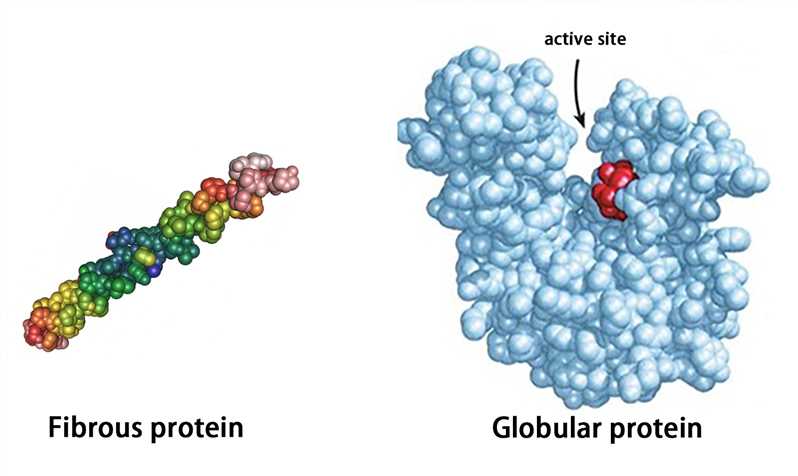 Figure 4. The representation of fibrous protein and globular protein.
Figure 4. The representation of fibrous protein and globular protein.
Quaternary Structure
Many proteins consist of a single polypeptide chain and have only three structural levels, however, some proteins are composed of multiple polypeptide chains. The spatial structure formed by the interaction of two or more polypeptide chains possessing independent tertiary structures (subunits) is called the quaternary structure. The subunits do not have to be covalently linked, but it is not excluded that some subunits are connected by disulfide bonds.
Complexes of multiple subunits are called multimers, and specifically, it would be called a dimer if it contains two subunits, a trimer if it contains three subunits, and so on. Additionally, multimers composed of identical subunits are referred to with the prefix “homo-” (e.g., a homodimer), and those composed of diverse subunits are referred to with the prefix "hetero-" (e.g., a heterodimer).
For multi-subunit proteins, the individual subunit has no biological activity, and only multimers with the intact quaternary structure are biologically active. Hemoglobin is a classic example of a protein with a quaternary structure. Hemoglobin is a tetramer formed by two alpha subunits and two beta subunits. The ability of hemoglobin to deliver oxygen to tissues depends on the binding of these subunits.
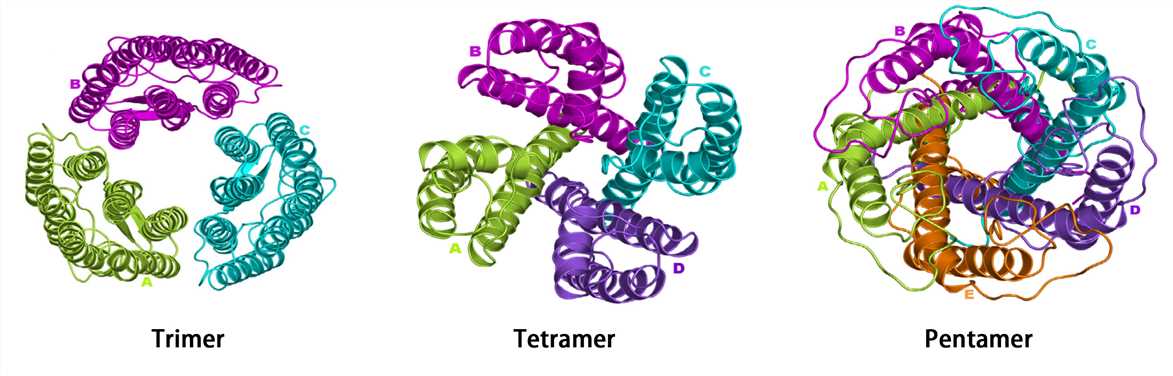 Figure 5. The quaternary structure.
Figure 5. The quaternary structure.
Creative Biostructure is specialized in the field of structural biology, and we provide contract services for the structural analysis of proteins of your interest.
References
- Sanger F, Thompson E O P. The amino-acid sequence in the glycyl chain of insulin. 1. The identification of lower peptides from partial hydrolysates. Biochemical Journal. 1953. 53(3): 353.
- Pauling L, et al. The structure of proteins: two hydrogen-bonded helical configurations of the polypeptide chain. Proceedings of the National Academy of Sciences. 1951. 37(4): 205-211.
- Chiang Y S, et al. New classification of supersecondary structures of sandwich‐like proteins uncovers strict patterns of strand assemblage. Proteins: Structure, Function, and Bioinformatics. 2007. 68(4): 915-921.
- Casciari D, et al. Quaternary structure predictions of transmembrane proteins starting from the monomer: a docking-based approach. BMC Bioinformatics. 2006. 7(1): 1-16.